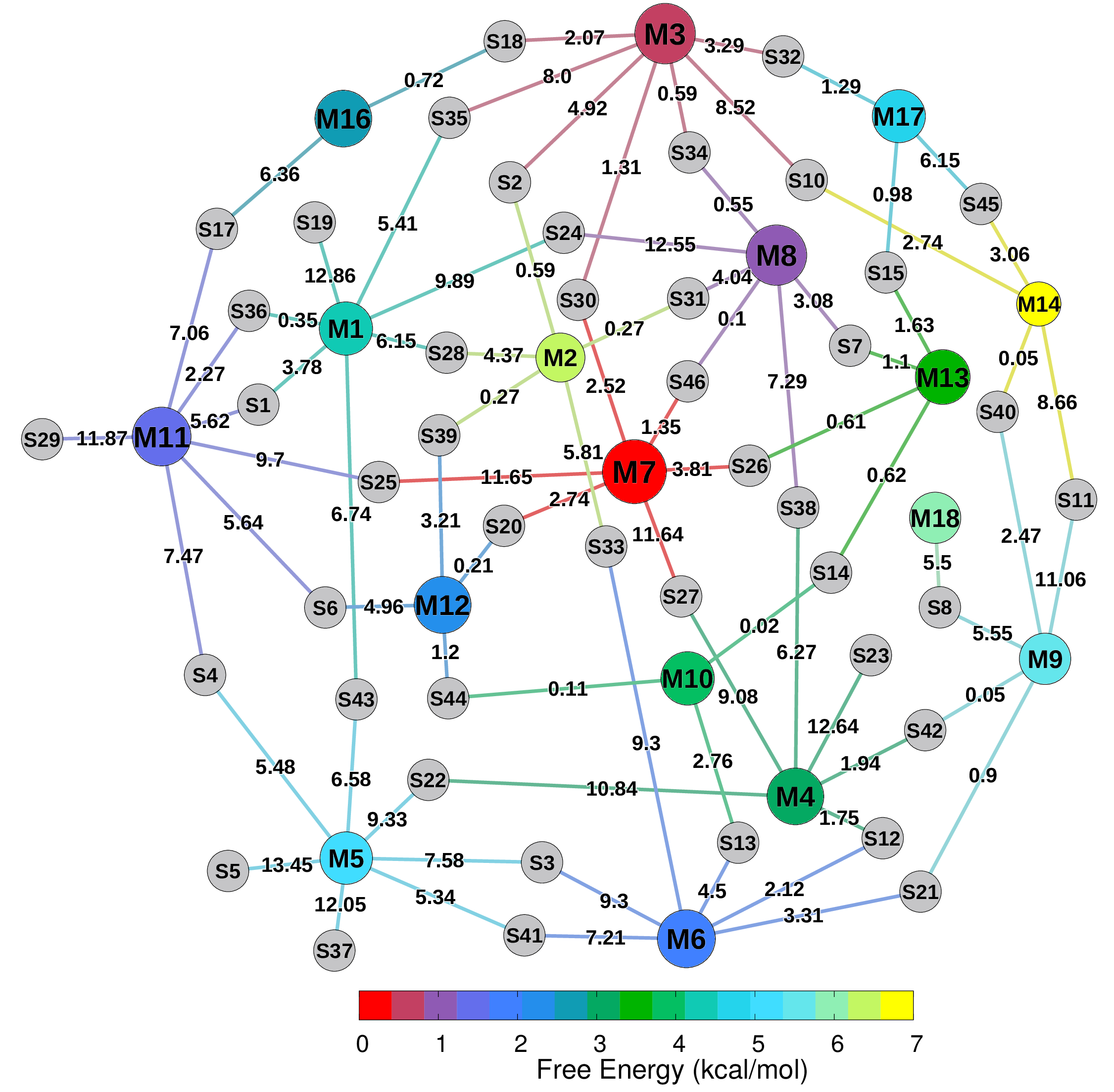
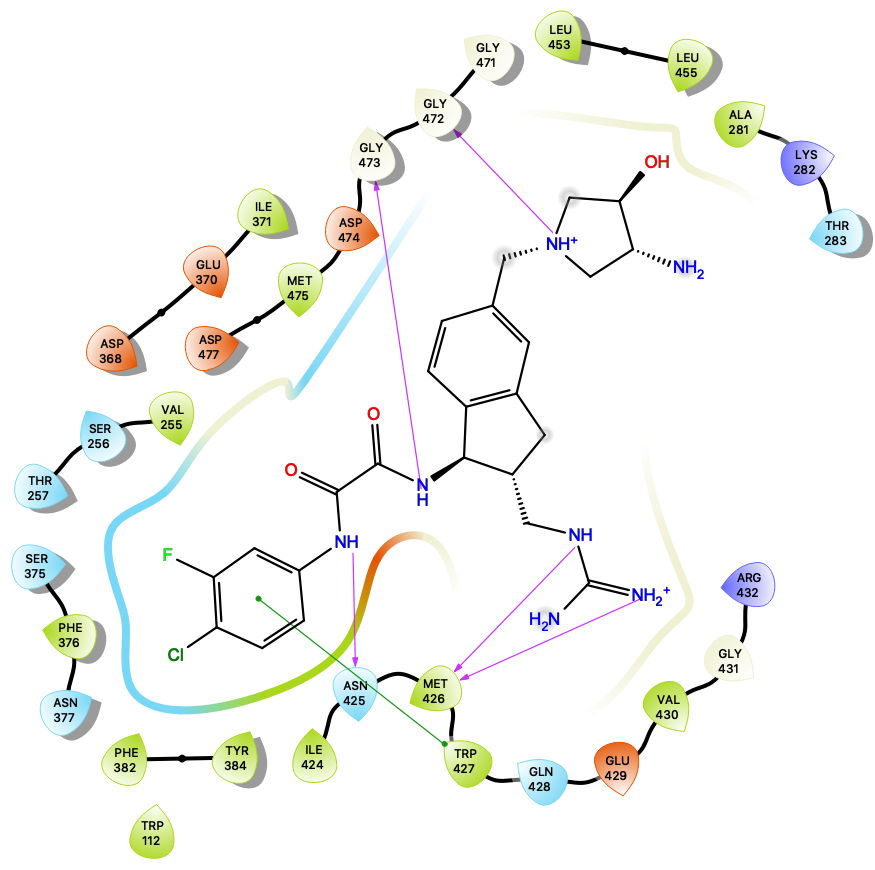

Welcome to the Molecular Simulations in Biology & Materials Group

Introducing Our CBE Faculty: Prof. Cameron Abrams
Recent News
Salman’s latest in Composites B
Congrats to Salman for publishing his latest paper in Composites B: Modeling sizing emulsion droplet ...
Read more
Ketan’s latest published in Soft Matter
Congrats to Ketan on the publication of his latest paper, “Atomistic Simulation of Volumetric Properties ...
Read more
Salman’s work accepted in Langmuir
Congrats to Salman for having his manuscript, “The Roles of Coupling Agent and Surfactant in ...
Read more
Steven Gossert’s work published in Protein Science
Congratulations to Steven for publishing his latest work on using molecular modeling to understand the ...
Read more