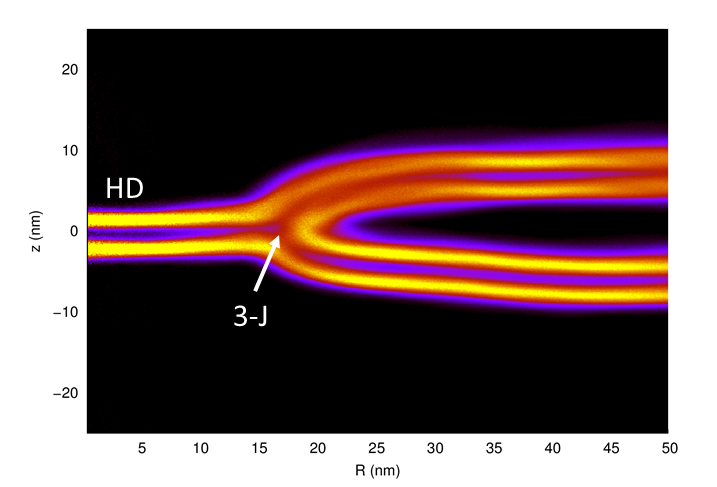
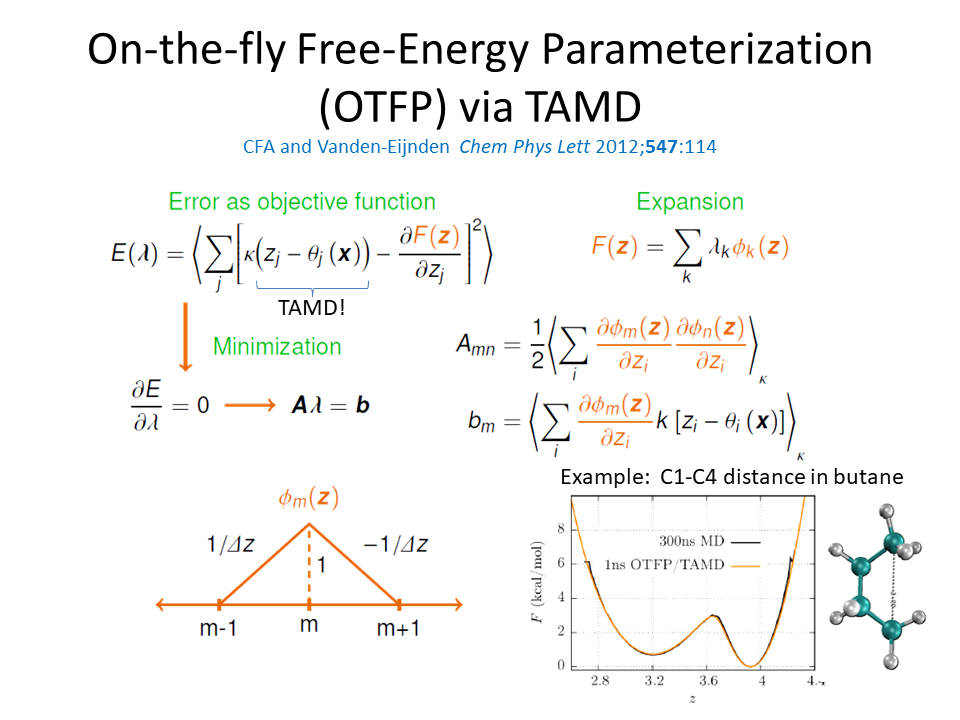
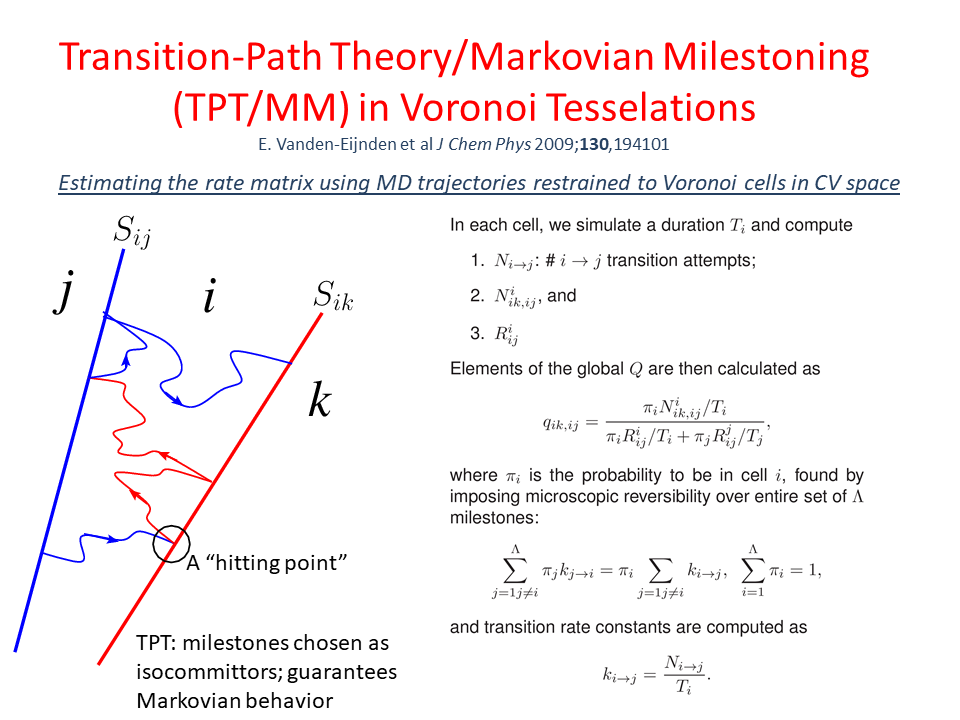
Molecular Simulations in Biology and Materials
The main expertise in the Abrams group is development and application of molecular dynamics (MD) simulations. We have particular interests in method development to enable enhanced sampling in all-atom MD, computational drug design, and simulations of polymeric materials and complex fluids. For the most up-to-date sampling of our research, please consult the publications page of this site.
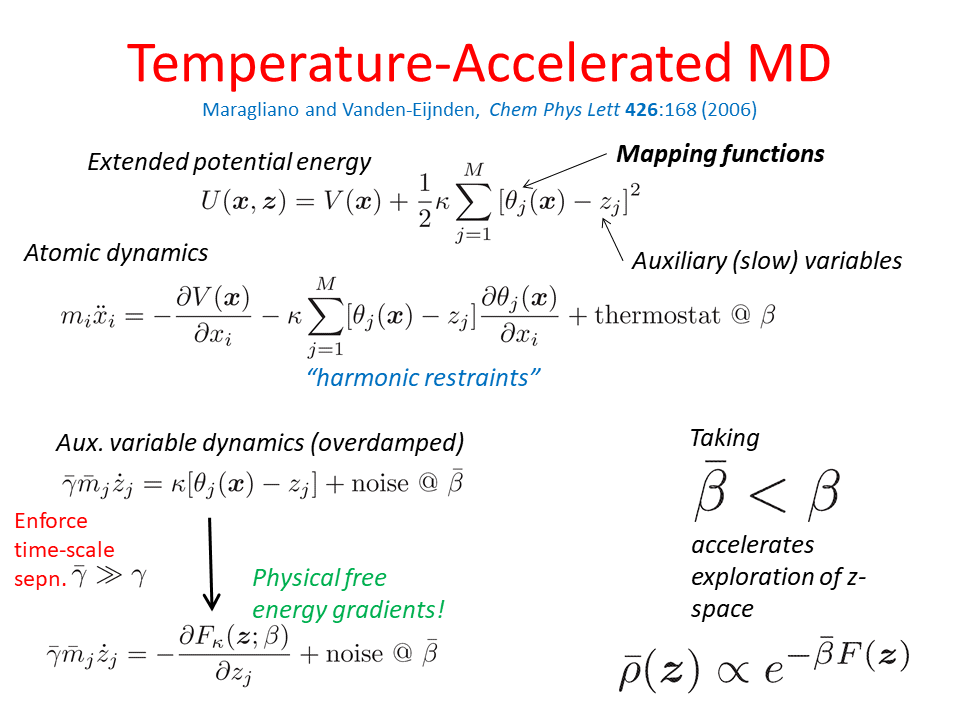
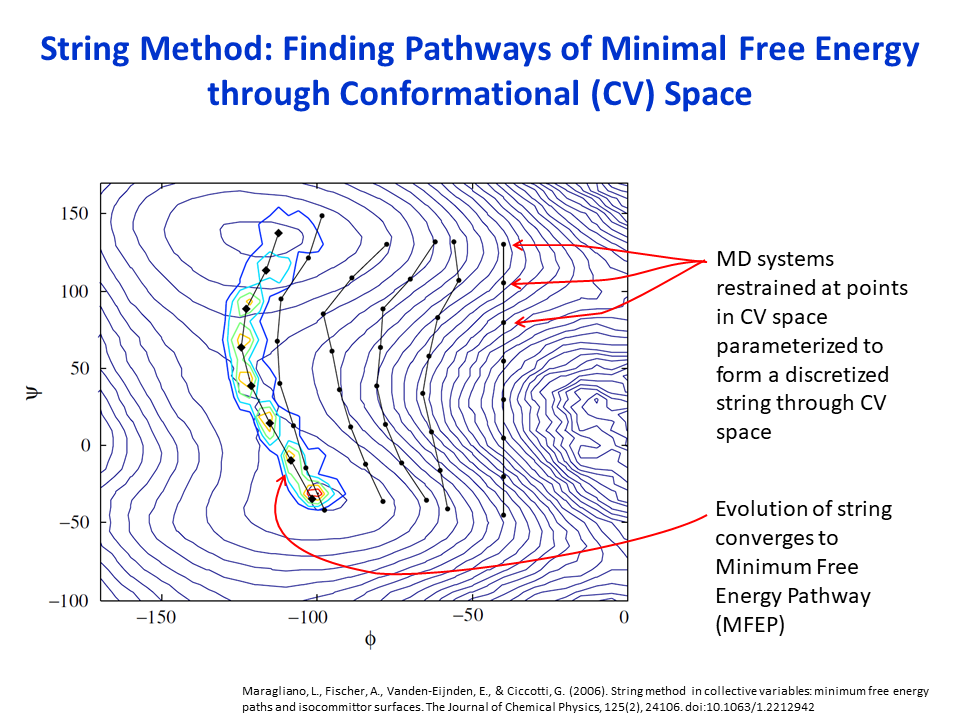
Enhanced Molecular Dynamics
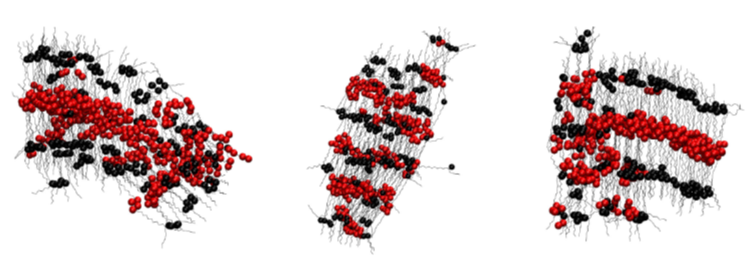
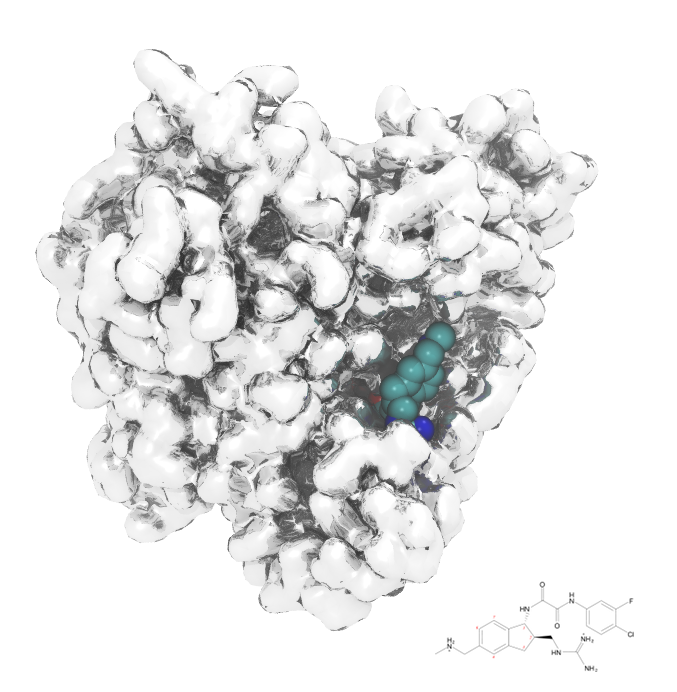
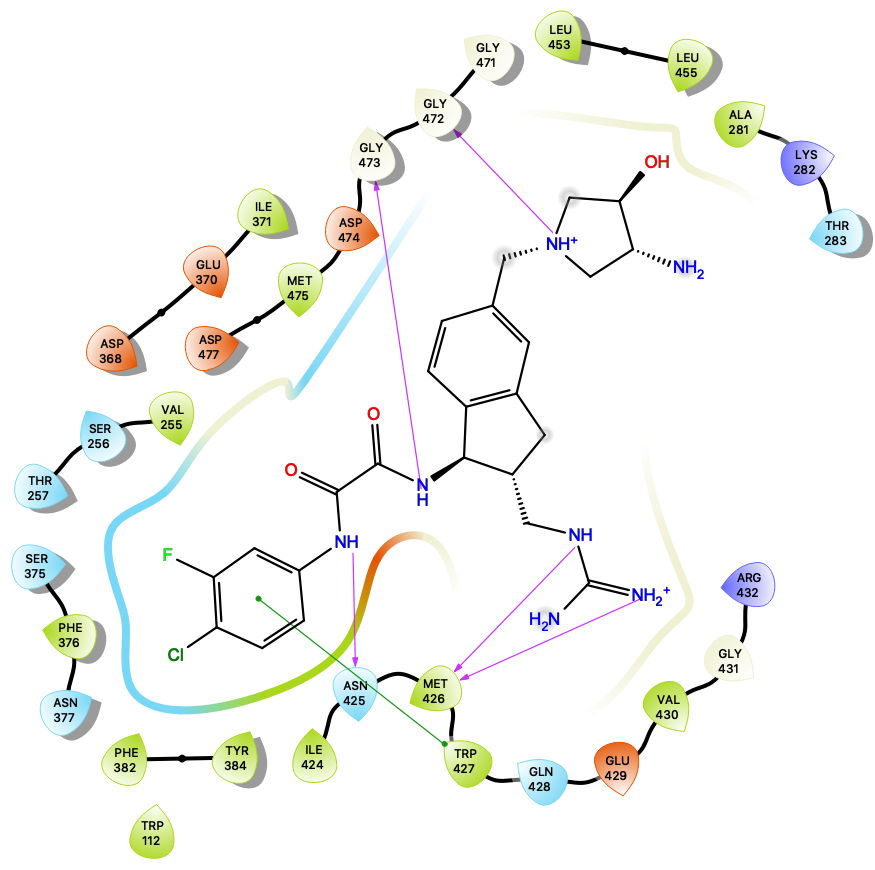
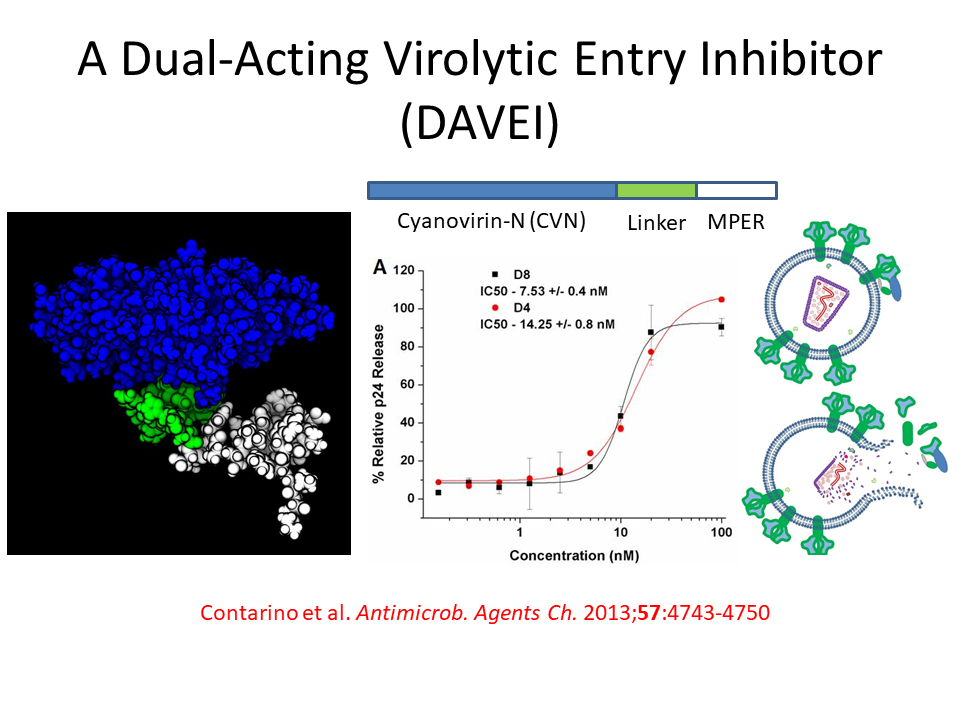
HIV-1 Fusion Machinery and its Antagonism
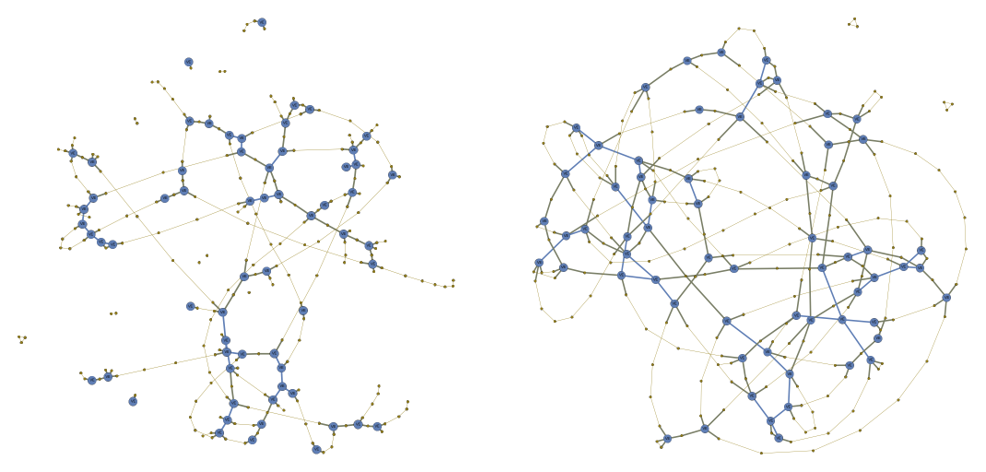
Thermoset Materials

Soft Matter