AFM and XRD#
%load_ext autoreload
%autoreload 2
import sys
import numpy as np
import matplotlib.pyplot as plt
sys.path.append('../../src/')
from m3_learning.nn.random import random_seed
from m3_learning.viz.style import set_style
from m3_learning.viz.printing import printer
from m3_learning.RHEED.AFM import visualize_afm_image, afm_substrate
from m3_learning.RHEED.XRD import plot_xrd, plot_rsm
from m3_learning.viz.layout import layout_fig, labelfigs
set_style("printing")
random_seed(seed=42)
2023-06-08 13:24:16.294663: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-06-08 13:24:16.847103: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:/home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:
2023-06-08 13:24:16.847170: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:/home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:
2023-06-08 13:24:16.847175: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
printing set for seaborn
Pytorch seed was set to 42
Numpy seed was set to 42
tensorflow seed was set to 42
Sample 1 - treated_213nm#
img2 = np.loadtxt('AFM/treated_213nm-film.txt')[:256]
scalebar_dict = {'image_size': 2008, 'scale_size': 500, 'units': 'nm'}
visualize_afm_image(img2, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict,)
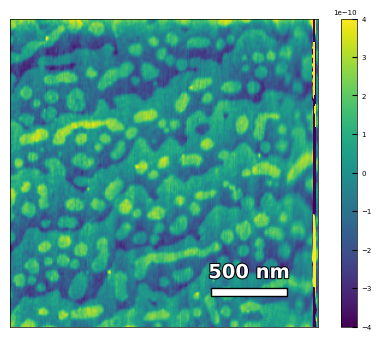
img3 = np.loadtxt('AFM/treated_213nm-substrate-tilted.txt')[:256]
analyzer = afm_substrate(img3, pixels=256, size=5e-6)
img_rot, size_rot = analyzer.rotate_image(angle=-50, demo=False)
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=60, prominence=1e-5, width=2, xz_angle=3, demo=False)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=3, prominence=1e-3, width=2, style='simple', fixed_height=3.91e-10, std_range=1, demo=False)
Step height = 3.91e-10 +- 0.00e+00
Step width = 2.13e-07 +- 8.87e-08
Miscut = 0.131° +- 0.074°
Sample 2 - treated_81nm:#
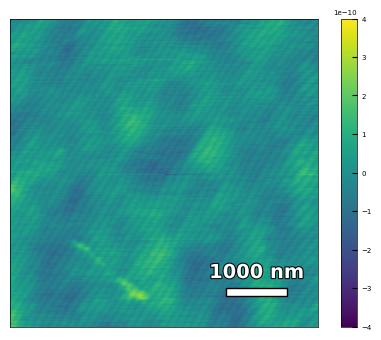
img1 = np.loadtxt('AFM/treated_81nm-substrate.txt')[:256]
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img1, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict)
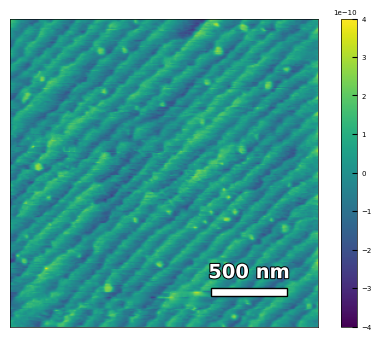
img2 = np.loadtxt('AFM/treated_81nm-film.txt')[:256]
scalebar_dict = {'image_size': 2008, 'scale_size': 500, 'units': 'nm'}
visualize_afm_image(img2, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict)
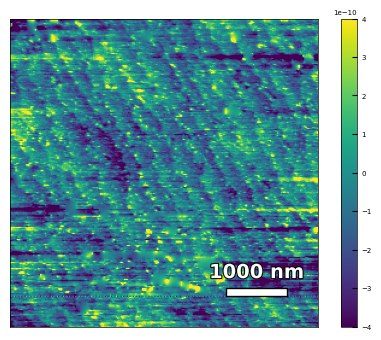
img3 = np.loadtxt('AFM/treated_81nm-substrate.txt')[:256]
analyzer = afm_substrate(img3, pixels=256, size=5e-6)
img_rot, size_rot = analyzer.rotate_image(angle=-56, demo=False)
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=60, prominence=1e-13, width=1.5, xz_angle=0, demo=False)
# step_heights, step_widths, miscut = analyzer.calculate_simple(x, z, peak_indices, fixed_height=3.91e-5, demo=False)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=0, prominence=1e-13, width=1.5,
style='simple', fixed_height=3.91e-10, std_range=1, demo=False)
Step height = 3.91e-10 +- 5.17e-26
Step width = 8.07e-08 +- 4.39e-08
Miscut = 0.330° +- 0.113°
Sample 3 - untreated_162nm#
img1 = np.loadtxt('AFM/untreated_162nm-substrate.txt')[:256]
img1 = np.rot90(img1, k=2)
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img1, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict)
img2 = np.loadtxt('AFM/untreated_162nm-film.txt')[:256]
scalebar_dict = {'image_size': 2008, 'scale_size': 500, 'units': 'nm'}
visualize_afm_image(img2, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict)
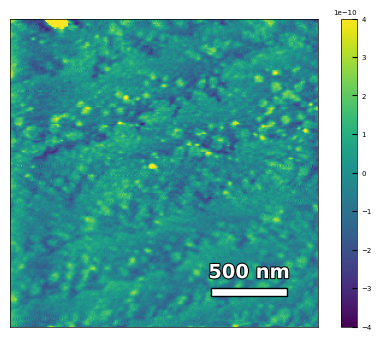
img3 = np.loadtxt('AFM/untreated_162nm-substrate-tilted.txt')[:256]
analyzer = afm_substrate(img3, pixels=256, size=5e-6)
img_rot, size_rot = analyzer.rotate_image(angle=65, demo=False)
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=60, prominence=1e-13, width=2, xz_angle=2, demo=False)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=2, prominence=1e-13, width=2, style='simple', fixed_height=3.91e-10/2, std_range=1, demo=False)
Step height = 1.95e-10 +- 0.00e+00
Step width = 1.62e-07 +- 8.27e-08
Miscut = 0.090° +- 0.135°
Summary of XRD and RSM#
fig = plt.figure(figsize=(8,10))
ax0 = plt.subplot2grid((4, 2), (0, 0), colspan=2) # colspan=2 means the plot spans 2 columns
files = ['./XRD/substrate-XRD_42_49.xrdml', './XRD/treated_213nm-XRD_42_29.xrdml', './XRD/treated_81nm-XRD_42_29.xrdml', './XRD/untreated_162nm-XRD_42_29.xrdml']
labels = ['substrate', 'treated_213nm', 'treated_81nm', 'untreated_162nm']
plot_xrd(ax0, files, labels, diff=None, xrange=(41.8, 49.2))
labelfigs(ax0, 0, loc='tr', size=15, style='b', inset_fraction=(0.8, 0.1))
files_002 = ['./XRD/treated_213nm-RSM_002.xrdml', './XRD/treated_81nm-RSM_002.xrdml', './XRD/untreated_162nm-RSM_002.xrdml']
for i, file in enumerate(files_002):
ax = plt.subplot2grid((4, 2), (i+1, 0))
plot_rsm(ax, file)
labelfigs(ax, i+1, loc='tr', size=15)
files_103 = ['./XRD/treated_213nm-RSM_103.xrdml', './XRD/treated_81nm-RSM_103.xrdml', './XRD/untreated_162nm-RSM_103.xrdml']
for i, file in enumerate(files_103):
ax = plt.subplot2grid((4, 2), (i+1, 1))
plot_rsm(ax, file)
labelfigs(ax, i+4, loc='tr', size=15)
plt.show()
print(f'\033[1mFig. S2 a\033[0m X-ray Diffraction result for a typical SrTiO3 substrate and samples. \
\033[1mb, c, d\033[0m Reciprocal Space Mapping results in (002) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively. \
\033[1me, f, g\033[0m Reciprocal Space Mapping results in (103) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively.')
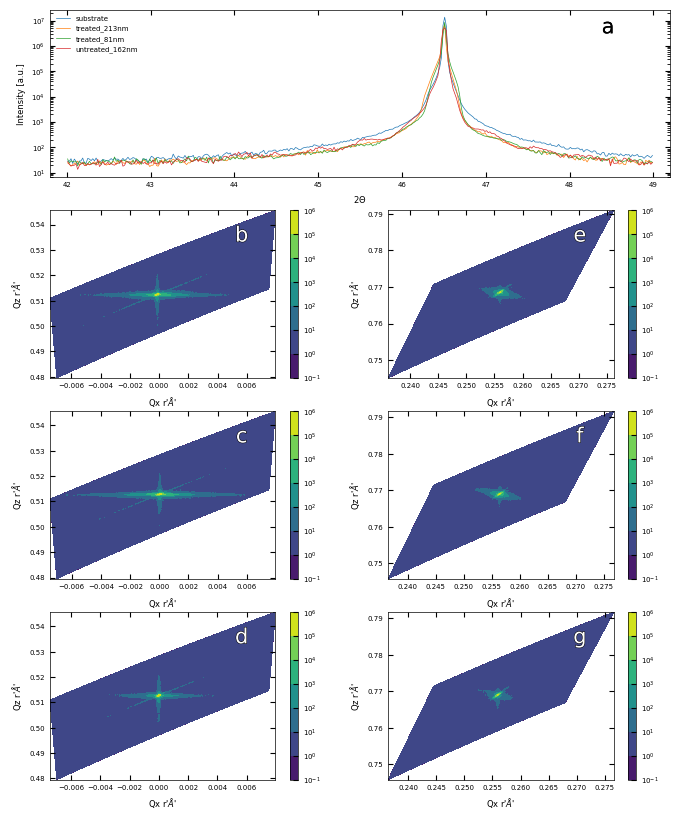
Fig. S2 a X-ray Diffraction result for a typical SrTiO3 substrate and samples. b, c, d Reciprocal Space Mapping results in (002) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively. e, f, g Reciprocal Space Mapping results in (103) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively.


