AFM and XRD with Details#
%load_ext autoreload
%autoreload 2
import os
import re
import numpy as np
import matplotlib.pyplot as plt
import sys
sys.path.append('../../src/')
from m3_learning.nn.random import random_seed
from m3_learning.viz.style import set_style
from m3_learning.util.file_IO import download_and_unzip
from m3_learning.viz.printing import printer
from m3_learning.viz.layout import layout_fig, labelfigs
from m3_learning.RHEED.AFM import visualize_afm_image, afm_substrate
from m3_learning.RHEED.XRD import plot_xrd, plot_rsm
# from m3_learning.be.dataset import BE_Dataset
printing = printer(basepath = 'Figures/2.AFM_XRD/')
set_style("printing")
random_seed(seed=42)
2023-06-08 14:43:01.392031: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-06-08 14:43:01.945151: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:/home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:
2023-06-08 14:43:01.945211: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory; LD_LIBRARY_PATH: /home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:/home/ferroelectric/micromamba/envs/m3-RHEED/lib/python3.10/site-packages/cv2/../../lib64:
2023-06-08 14:43:01.945215: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
printing set for seaborn
Pytorch seed was set to 42
Numpy seed was set to 42
tensorflow seed was set to 42
1. Download datasets from Zenodo#
# Download the data file from Zenodo
download=False
if download:
# full size version
urls = ['https://zenodo.org/record/8000271/files/AFM.zip?download=1',
'https://zenodo.org/record/8000271/files/XRD.zip?download=1']
for url in urls:
# Specify the filename and the path to save the file
filename = re.split(r'\?', os.path.basename(url))[0]
save_path = './../2023_RHEED_PLD_SrTiO3/'
# download the file
download_and_unzip(filename, url, save_path)
if os.path.isfile('./../2023_RHEED_PLD_SrTiO3/AFM.zip'): os.remove('./../2023_RHEED_PLD_SrTiO3/AFM.zip')
if os.path.isfile('./../2023_RHEED_PLD_SrTiO3/XRD.zip'): os.remove('./../2023_RHEED_PLD_SrTiO3/XRD.zip')
2. AFM Results Analysis#
2.1 Sample 1 - treated_213nm#
2.1.1 Visualize and save the afm figures with colorbar and scalebar#
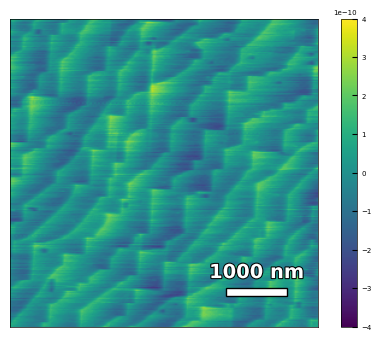
img1 = np.loadtxt('AFM/treated_213nm-substrate.txt')[:256]
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img1, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict,
filename='treated_213nm-substrate', printing=printing)
Figures/2.AFM_XRD/treated_213nm-substrate.png
Figures/2.AFM_XRD/treated_213nm-substrate.svg
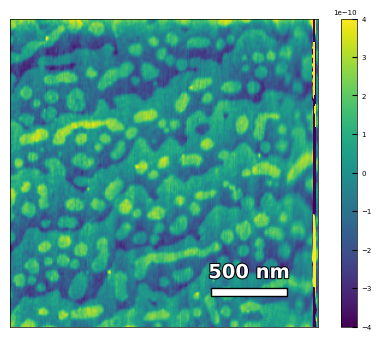
img2 = np.loadtxt('AFM/treated_213nm-film.txt')[:256]
scalebar_dict = {'image_size': 2008, 'scale_size': 500, 'units': 'nm'}
visualize_afm_image(img2, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict,
filename='treated_213nm-film', printing=printing)
Figures/2.AFM_XRD/treated_213nm-film.png
Figures/2.AFM_XRD/treated_213nm-film.svg
2.1.2 Calculate the substrate step height, step width, and miscut#
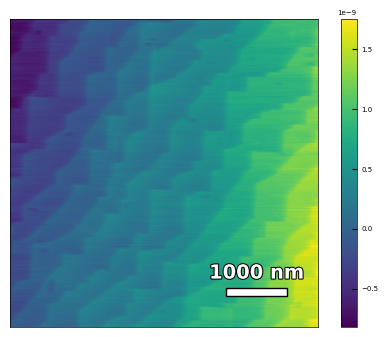
img3 = np.loadtxt('AFM/treated_213nm-substrate-tilted.txt')[:256]
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img3, colorbar_range=None, figsize=(6,4), scalebar_dict=scalebar_dict)
# rotate the angle to make step edges horizontal
analyzer = afm_substrate(img3, pixels=256, size=5e-6)
img_rot, size_rot = analyzer.rotate_image(angle=-50)
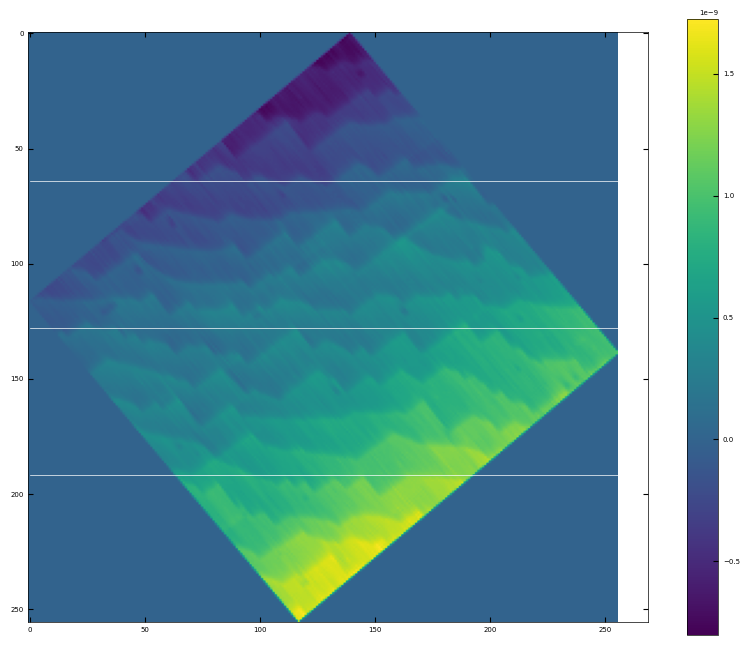
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=60, prominence=1e-5, width=2, xz_angle=3, demo=True)
# with the provided height from literature
step_heights, step_widths, miscut = analyzer.calculate_simple(x, z, peak_indices, fixed_height=3.91e-10, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=3, prominence=1e-3, width=2, style='simple', fixed_height=3.91e-10, std_range=1, demo=False)
Step 1: Height = 3.91e-10, Width = 1.62e-07, Miscut = 0.138°
Step 2: Height = 3.91e-10, Width = 1.40e-07, Miscut = 0.160°
Step 3: Height = 3.91e-10, Width = 1.08e-07, Miscut = 0.208°
Step 4: Height = 3.91e-10, Width = 9.57e-08, Miscut = 0.234°
Step 5: Height = 3.91e-10, Width = 1.94e-07, Miscut = 0.115°
Step 6: Height = 3.91e-10, Width = 1.71e-07, Miscut = 0.131°
Step 7: Height = 3.91e-10, Width = 3.99e-08, Miscut = 0.561°
Step 8: Height = 3.91e-10, Width = 1.23e-07, Miscut = 0.183°
Step 9: Height = 3.91e-10, Width = 2.11e-07, Miscut = 0.106°
Results:
Average step height = 3.91e-10, Standard deviation = 0.00e+00
Average step width = 1.38e-07, Standard deviation = 5.03e-08
Average miscut = 0.204°, Standard deviation = 0.132°
Step height = 3.91e-10 +- 0.00e+00
Step width = 2.13e-07 +- 8.87e-08
Miscut = 0.131° +- 0.074°
# calculate the height from afm figure
step_heights, step_widths, miscut = analyzer.calculate_simple(x, z, peak_indices, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=3, prominence=1e-10, width=2, style='simple', std_range=1, demo=False)
Step 1: Height = 1.73e-10, Width = 1.62e-07, Miscut = 0.061°
Step 2: Height = 1.21e-10, Width = 1.40e-07, Miscut = 0.050°
Step 3: Height = -1.00e-10, Width = 1.08e-07, Miscut = -0.053°
Step 4: Height = 1.45e-10, Width = 9.57e-08, Miscut = 0.087°
Step 5: Height = 1.32e-10, Width = 1.94e-07, Miscut = 0.039°
Step 6: Height = 8.74e-11, Width = 1.71e-07, Miscut = 0.029°
Step 7: Height = 1.63e-11, Width = 3.99e-08, Miscut = 0.023°
Step 8: Height = 2.25e-11, Width = 1.23e-07, Miscut = 0.011°
Step 9: Height = 1.05e-10, Width = 2.11e-07, Miscut = 0.029°
Results:
Average step height = 7.79e-11, Standard deviation = 8.01e-11
Average step width = 1.38e-07, Standard deviation = 5.03e-08
Average miscut = 0.031°, Standard deviation = 0.037°
Step height = 1.50e-10 +- 1.28e-10
Step width = 2.13e-07 +- 8.87e-08
Miscut = 0.042° +- 0.038°
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=100, prominence=1e-11, width=2, xz_angle=0, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_fit(x, z, peak_indices, valley_indices, fixed_height=3.91e-10, demo=True)
step 1: step_width: 1.10e-10, left_peak: (4.99e-07, -4.60e-10), valley: (5.41e-07, -5.11e-10), right_peak: (6.93e-07, -2.94e-10)
step 2: step_width: 4.97e-10, left_peak: (6.93e-07, -2.94e-10), valley: (8.18e-07, -3.74e-10), right_peak: (8.87e-07, -1.42e-10)
step 3: step_width: 2.44e-10, left_peak: (9.57e-07, -1.84e-10), valley: (1.03e-06, -2.97e-10), right_peak: (1.15e-06, -6.09e-11)
step 4: step_width: 2.04e-10, left_peak: (1.33e-06, 1.19e-10), valley: (1.40e-06, 2.98e-11), right_peak: (1.52e-06, 2.37e-10)
step 5: step_width: 2.84e-10, left_peak: (1.52e-06, 2.37e-10), valley: (1.58e-06, 1.46e-10), right_peak: (1.65e-06, 3.88e-10)
step 6: step_width: 3.97e-10, left_peak: (1.73e-06, 2.83e-10), valley: (1.87e-06, 1.73e-10), right_peak: (1.97e-06, 3.74e-10)
step 7: step_width: 5.13e-10, left_peak: (1.97e-06, 3.74e-10), valley: (2.11e-06, 1.85e-10), right_peak: (2.19e-06, 3.79e-10)
step 8: step_width: 2.68e-10, left_peak: (2.19e-06, 3.79e-10), valley: (2.26e-06, 3.16e-10), right_peak: (2.33e-06, 5.22e-10)
step 9: step_width: 4.10e-10, left_peak: (2.33e-06, 5.22e-10), valley: (2.41e-06, 4.52e-10), right_peak: (2.47e-06, 6.79e-10)
step 10: step_width: 4.29e-10, left_peak: (2.47e-06, 6.79e-10), valley: (2.63e-06, 5.62e-10), right_peak: (2.76e-06, 7.97e-10)
step 11: step_width: 2.41e-10, left_peak: (2.76e-06, 7.97e-10), valley: (2.81e-06, 7.03e-10), right_peak: (2.91e-06, 9.61e-10)
step 12: step_width: 5.79e-11, left_peak: (2.91e-06, 9.61e-10), valley: (2.94e-06, 9.25e-10), right_peak: (2.97e-06, 9.47e-10)
Results:
Average step height = 3.91e-10, Standard deviation = 0.00e+00
Average step width = 3.05e-10, Standard deviation = 1.40e-10
Average miscut = 54.187°, Standard deviation = 13.357°
The “simple” method with provided step height gives a more accurate results for the step width and miscut:
Step height = 3.91e-10 +- 0.00e+00
Step width = 2.13e-07 +- 8.87e-08
Miscut = 0.131° +- 0.074°
2.2 Sample 2 - treated_81nm:#
2.2.1 Visualize and save the afm figures with colorbar and scalebar#
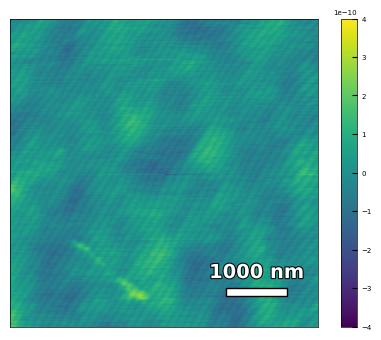
img1 = np.loadtxt('AFM/treated_81nm-substrate.txt')[:256]
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img1, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict,
filename='treated_81nm-substrate', printing=printing)
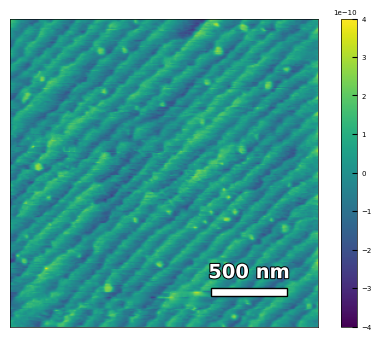
img2 = np.loadtxt('AFM/treated_81nm-film.txt')[:256]
scalebar_dict = {'image_size': 2008, 'scale_size': 500, 'units': 'nm'}
visualize_afm_image(img2, colorbar_range=[-4e-10, 4e-10], figsize=(6,4), scalebar_dict=scalebar_dict,
filename='treated_81nm-film', printing=printing)
Figures/2.AFM_XRD/treated_81nm-substrate.png
Figures/2.AFM_XRD/treated_81nm-substrate.svg
Figures/2.AFM_XRD/treated_81nm-film.png
Figures/2.AFM_XRD/treated_81nm-film.svg
2.2.2 Calculate the substrate step height, step width, and miscut#
img3 = np.loadtxt('AFM/treated_81nm-substrate.txt')[:256]
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img3, colorbar_range=None, figsize=(6,4), scalebar_dict=scalebar_dict)
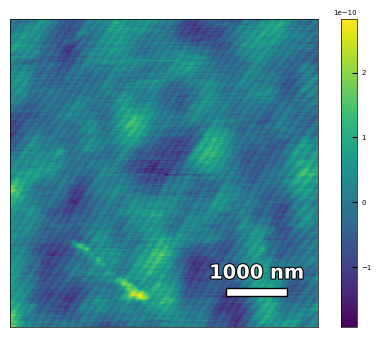
# rotate the angle to make step edges horizontal
analyzer = afm_substrate(img3, pixels=256, size=5e-6)
img_rot, size_rot = analyzer.rotate_image(angle=-56)
prominence = 1e-13
width = 1.5
xz_angle = 0
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=60, prominence=prominence, width=width, xz_angle=xz_angle, demo=True)
# with the provided height from literature
step_heights, step_widths, miscut = analyzer.calculate_simple(x, z, peak_indices, fixed_height=3.91e-5, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=xz_angle, prominence=prominence, width=width,
style='simple', fixed_height=3.91e-10, std_range=1, demo=False)
Step 1: Height = 3.91e-05, Width = 1.69e-07, Miscut = 89.753°
Step 2: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 3: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 4: Height = 3.91e-05, Width = 9.85e-08, Miscut = 89.856°
Step 5: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 6: Height = 3.91e-05, Width = 1.13e-07, Miscut = 89.835°
Step 7: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 8: Height = 3.91e-05, Width = 8.44e-08, Miscut = 89.876°
Step 9: Height = 3.91e-05, Width = 1.13e-07, Miscut = 89.835°
Step 10: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 11: Height = 3.91e-05, Width = 1.27e-07, Miscut = 89.814°
Step 12: Height = 3.91e-05, Width = 1.69e-07, Miscut = 89.753°
Step 13: Height = 3.91e-05, Width = 1.69e-07, Miscut = 89.753°
Step 14: Height = 3.91e-05, Width = 1.27e-07, Miscut = 89.814°
Step 15: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 16: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 17: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 18: Height = 3.91e-05, Width = 5.63e-08, Miscut = 89.918°
Step 19: Height = 3.91e-05, Width = 1.13e-07, Miscut = 89.835°
Results:
Average step height = 3.91e-05, Standard deviation = 6.78e-21
Average step width = 9.40e-08, Standard deviation = 4.16e-08
Average miscut = 89.862°, Standard deviation = 0.061°
Step height = 3.91e-10 +- 5.17e-26
Step width = 8.07e-08 +- 4.39e-08
Miscut = 0.330° +- 0.113°
# calculate the height from afm figure
step_heights, step_widths, miscut = analyzer.calculate_simple(x, z, peak_indices, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=2, prominence=1e-2, width=1.5, style='simple', std_range=1, demo=False)
Step 1: Height = -4.61e-11, Width = 1.69e-07, Miscut = -0.016°
Step 2: Height = -1.44e-11, Width = 5.63e-08, Miscut = -0.015°
Step 3: Height = -2.18e-11, Width = 5.63e-08, Miscut = -0.022°
Step 4: Height = -3.33e-11, Width = 9.85e-08, Miscut = -0.019°
Step 5: Height = -6.26e-12, Width = 5.63e-08, Miscut = -0.006°
Step 6: Height = -2.23e-11, Width = 1.13e-07, Miscut = -0.011°
Step 7: Height = 1.97e-11, Width = 5.63e-08, Miscut = 0.020°
Step 8: Height = 3.62e-11, Width = 8.44e-08, Miscut = 0.025°
Step 9: Height = 1.46e-11, Width = 1.13e-07, Miscut = 0.007°
Step 10: Height = 5.60e-11, Width = 5.63e-08, Miscut = 0.057°
Step 11: Height = 1.11e-11, Width = 1.27e-07, Miscut = 0.005°
Step 12: Height = 1.05e-10, Width = 1.69e-07, Miscut = 0.036°
Step 13: Height = 6.57e-11, Width = 1.69e-07, Miscut = 0.022°
Step 14: Height = -1.67e-10, Width = 1.27e-07, Miscut = -0.076°
Step 15: Height = -4.48e-12, Width = 5.63e-08, Miscut = -0.005°
Step 16: Height = -1.28e-12, Width = 5.63e-08, Miscut = -0.001°
Step 17: Height = -1.74e-11, Width = 5.63e-08, Miscut = -0.018°
Step 18: Height = -2.77e-11, Width = 5.63e-08, Miscut = -0.028°
Step 19: Height = 3.06e-12, Width = 1.13e-07, Miscut = 0.002°
Results:
Average step height = -2.65e-12, Standard deviation = 5.32e-11
Average step width = 9.40e-08, Standard deviation = 4.16e-08
Average miscut = -0.002°, Standard deviation = 0.027°
Step height = 3.53e-13 +- 3.97e-11
Step width = 8.05e-08 +- 5.22e-08
Miscut = 0.008° +- 0.031°
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=100, prominence=1e-13, width=1, xz_angle=0, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_fit(x, z, peak_indices, valley_indices, fixed_height=3.91e-10, demo=True)
step 1: step_width: 4.33e-11, left_peak: (5.35e-07, -7.14e-11), valley: (5.49e-07, -9.73e-11), right_peak: (5.91e-07, -4.48e-11)
step 2: step_width: 3.45e-11, left_peak: (6.75e-07, 7.57e-12), valley: (6.89e-07, -8.11e-12), right_peak: (7.46e-07, 6.73e-11)
step 3: step_width: 1.00e-10, left_peak: (7.46e-07, 6.73e-11), valley: (7.74e-07, 1.30e-11), right_peak: (7.88e-07, 3.60e-11)
step 4: step_width: 6.81e-11, left_peak: (7.88e-07, 3.60e-11), valley: (8.16e-07, -4.68e-12), right_peak: (8.44e-07, 2.27e-11)
step 5: step_width: 7.35e-11, left_peak: (8.44e-07, 2.27e-11), valley: (8.72e-07, -1.13e-11), right_peak: (9.14e-07, 4.78e-11)
step 6: step_width: 4.25e-11, left_peak: (9.14e-07, 4.78e-11), valley: (9.29e-07, 2.03e-11), right_peak: (9.71e-07, 6.54e-11)
step 7: step_width: 6.54e-11, left_peak: (9.71e-07, 6.54e-11), valley: (9.99e-07, 1.68e-11), right_peak: (1.03e-06, 3.35e-11)
step 10: step_width: 1.13e-10, left_peak: (1.17e-06, -4.47e-12), valley: (1.21e-06, -4.84e-11), right_peak: (1.24e-06, -2.20e-12)
step 11: step_width: 1.05e-10, left_peak: (1.24e-06, -2.20e-12), valley: (1.27e-06, -6.07e-11), right_peak: (1.29e-06, -1.44e-11)
step 12: step_width: 7.03e-11, left_peak: (1.29e-06, -1.44e-11), valley: (1.32e-06, -4.34e-11), right_peak: (1.36e-06, 1.86e-11)
step 13: step_width: 6.72e-11, left_peak: (1.36e-06, 1.86e-11), valley: (1.39e-06, -1.50e-11), right_peak: (1.44e-06, 3.55e-11)
step 14: step_width: 6.50e-11, left_peak: (1.44e-06, 3.55e-11), valley: (1.45e-06, -1.48e-11), right_peak: (1.49e-06, 2.91e-11)
step 15: step_width: 4.30e-11, left_peak: (1.49e-06, 2.91e-11), valley: (1.51e-06, 7.15e-12), right_peak: (1.53e-06, 4.92e-11)
step 16: step_width: 7.76e-11, left_peak: (1.53e-06, 4.92e-11), valley: (1.58e-06, -1.30e-11), right_peak: (1.60e-06, -2.78e-12)
step 17: step_width: 2.38e-11, left_peak: (1.60e-06, -2.78e-12), valley: (1.62e-06, -1.16e-11), right_peak: (1.66e-06, 3.35e-11)
step 18: step_width: 8.43e-11, left_peak: (1.66e-06, 3.35e-11), valley: (1.69e-06, -4.80e-12), right_peak: (1.73e-06, 6.42e-11)
step 19: step_width: 5.16e-11, left_peak: (1.73e-06, 6.42e-11), valley: (1.76e-06, 4.10e-11), right_peak: (1.79e-06, 6.94e-11)
step 20: step_width: 5.74e-11, left_peak: (1.79e-06, 6.94e-11), valley: (1.81e-06, 4.28e-11), right_peak: (1.84e-06, 7.36e-11)
step 21: step_width: 1.08e-10, left_peak: (1.84e-06, 7.36e-11), valley: (1.89e-06, 1.93e-11), right_peak: (1.91e-06, 5.54e-11)
step 22: step_width: 7.02e-11, left_peak: (1.94e-06, 6.42e-11), valley: (2.00e-06, 3.07e-11), right_peak: (2.03e-06, 4.91e-11)
step 23: step_width: 7.65e-11, left_peak: (2.03e-06, 4.91e-11), valley: (2.05e-06, 2.29e-11), right_peak: (2.07e-06, 4.80e-11)
step 24: step_width: 6.56e-11, left_peak: (2.07e-06, 4.80e-11), valley: (2.11e-06, 1.31e-11), right_peak: (2.14e-06, 3.36e-11)
step 25: step_width: 7.12e-11, left_peak: (2.14e-06, 3.36e-11), valley: (2.17e-06, 1.70e-11), right_peak: (2.19e-06, 7.16e-11)
step 26: step_width: 1.53e-10, left_peak: (2.19e-06, 7.16e-11), valley: (2.24e-06, 1.91e-11), right_peak: (2.25e-06, 5.26e-11)
step 27: step_width: 8.46e-11, left_peak: (2.25e-06, 5.26e-11), valley: (2.28e-06, -1.28e-11), right_peak: (2.31e-06, 6.36e-12)
step 28: step_width: 1.33e-10, left_peak: (2.35e-06, -3.54e-11), valley: (2.42e-06, -8.75e-11), right_peak: (2.45e-06, -5.49e-11)
step 29: step_width: 8.35e-11, left_peak: (2.45e-06, -5.49e-11), valley: (2.48e-06, -1.07e-10), right_peak: (2.50e-06, -7.59e-11)
step 30: step_width: 7.23e-11, left_peak: (2.50e-06, -7.59e-11), valley: (2.53e-06, -1.03e-10), right_peak: (2.56e-06, -5.79e-11)
step 31: step_width: 8.29e-11, left_peak: (2.56e-06, -5.79e-11), valley: (2.60e-06, -7.78e-11), right_peak: (2.65e-06, -1.48e-11)
step 32: step_width: 2.00e-10, left_peak: (2.65e-06, -1.48e-11), valley: (2.72e-06, -7.31e-11), right_peak: (2.76e-06, 1.16e-11)
step 33: step_width: 8.06e-11, left_peak: (2.76e-06, 1.16e-11), valley: (2.79e-06, -4.12e-11), right_peak: (2.83e-06, 3.85e-13)
step 34: step_width: 6.10e-11, left_peak: (2.83e-06, 3.85e-13), valley: (2.84e-06, -3.85e-11), right_peak: (2.88e-06, 2.77e-11)
step 35: step_width: 9.80e-11, left_peak: (2.88e-06, 2.77e-11), valley: (2.91e-06, -3.05e-12), right_peak: (2.94e-06, 6.43e-11)
step 36: step_width: 7.45e-11, left_peak: (2.94e-06, 6.43e-11), valley: (2.97e-06, 3.25e-11), right_peak: (3.00e-06, 7.53e-11)
step 37: step_width: 1.10e-10, left_peak: (3.00e-06, 7.53e-11), valley: (3.04e-06, 4.39e-11), right_peak: (3.08e-06, 1.23e-10)
step 38: step_width: 6.71e-11, left_peak: (3.08e-06, 1.23e-10), valley: (3.10e-06, 7.49e-11), right_peak: (3.12e-06, 1.13e-10)
step 39: step_width: 1.10e-10, left_peak: (3.12e-06, 1.13e-10), valley: (3.15e-06, 5.41e-11), right_peak: (3.18e-06, 1.05e-10)
step 40: step_width: 1.91e-10, left_peak: (3.18e-06, 1.05e-10), valley: (3.26e-06, -1.77e-11), right_peak: (3.29e-06, 5.28e-12)
step 41: step_width: 4.12e-11, left_peak: (3.29e-06, 5.28e-12), valley: (3.31e-06, -1.66e-11), right_peak: (3.35e-06, 4.16e-11)
step 42: step_width: 7.53e-11, left_peak: (3.35e-06, 4.16e-11), valley: (3.38e-06, 6.47e-12), right_peak: (3.42e-06, 6.67e-11)
Results:
Average step height = 3.91e-10, Standard deviation = 0.00e+00
Average step width = 8.24e-11, Standard deviation = 3.67e-11
Average miscut = 78.209°, Standard deviation = 4.957°
# with the provided height from literature
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=0, prominence=1e-13, width=1, style='fit', fixed_height=3.91e-10, std_range=1, demo=False)
Step height = 3.91e-10 +- 0.00e+00
Step width = 8.55e-11 +- 4.62e-11
Miscut = 77.852° +- 5.976°
The “simple” method with provided step height gives a more accurate results for the step width and miscut:
Step height = 3.91e-10 +- 5.17e-26
Step width = 8.07e-08 +- 4.39e-08
Miscut = 0.330° +- 0.113°
2.3 Sample 3 - untreated_162nm#
2.3.1 Visualize and save the afm figures with colorbar and scalebar#
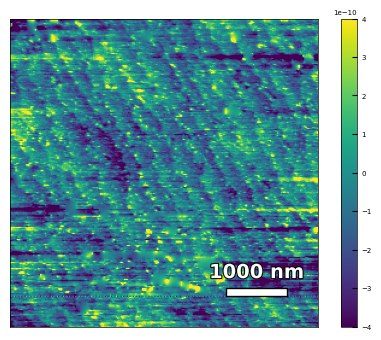
img1 = np.loadtxt('AFM/untreated_162nm-substrate.txt')[:256]
img1 = np.rot90(img1, k=2)
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img1, colorbar_range=[-4e-10, 4e-10], figsize=(6,4),
scalebar_dict=scalebar_dict, filename='untreated_162nm-substrate', printing=printing)
Figures/2.AFM_XRD/untreated_162nm-substrate.png
Figures/2.AFM_XRD/untreated_162nm-substrate.svg
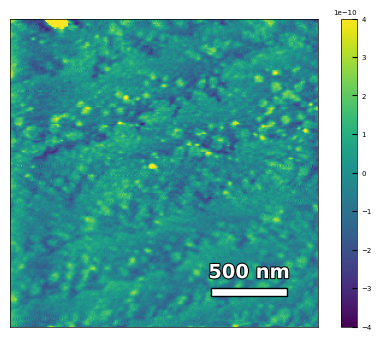
img2 = np.loadtxt('AFM/untreated_162nm-film.txt')[:256]
scalebar_dict = {'image_size': 2008, 'scale_size': 500, 'units': 'nm'}
visualize_afm_image(img2, colorbar_range=[-4e-10, 4e-10], figsize=(6,4),
scalebar_dict=scalebar_dict, filename='untreated_162nm-film', printing=printing)
Figures/2.AFM_XRD/untreated_162nm-film.png
Figures/2.AFM_XRD/untreated_162nm-film.svg
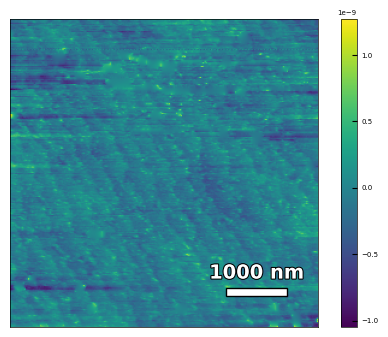
2.3.2 Calculates the substrate step height, step width, and miscut#
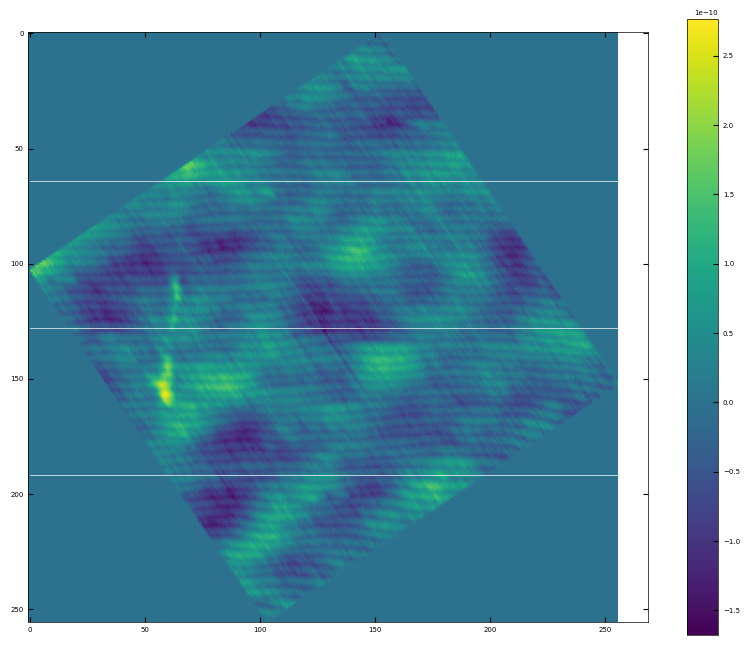
img3 = np.loadtxt('AFM/untreated_162nm-substrate-tilted.txt')[:256]
scalebar_dict = {'image_size': 5000, 'scale_size': 1000, 'units': 'nm'}
visualize_afm_image(img3, colorbar_range=None, figsize=(6,4), scalebar_dict=scalebar_dict)
# rotate the angle to make step edges horizontal
analyzer = afm_substrate(img3, pixels=256, size=5e-6)
img_rot, size_rot = analyzer.rotate_image(angle=65)
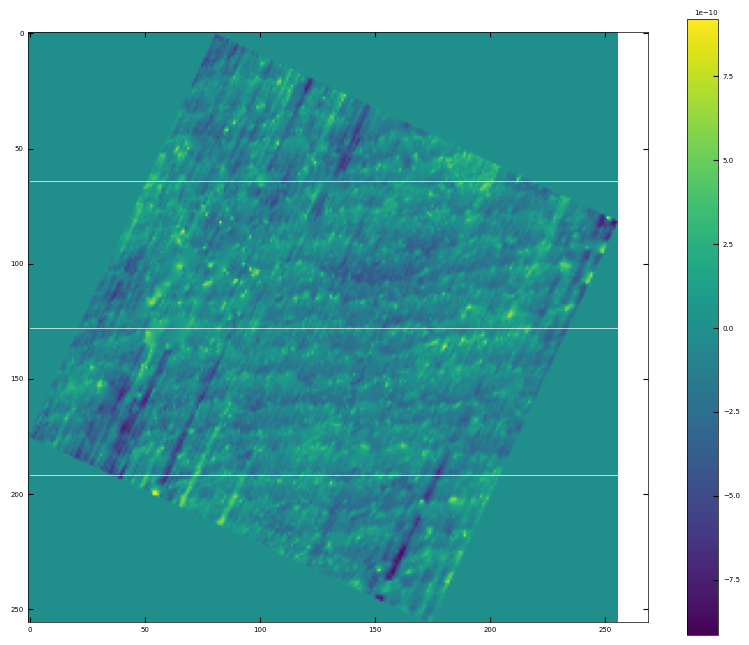
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=60, prominence=1e-13, width=2, xz_angle=2, demo=True)
# with the provided height from literature
step_heights, step_widths, miscut = analyzer.calculate_simple(x, z, peak_indices, fixed_height=3.91e-10/2, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=2, prominence=1e-13, width=2, style='simple', fixed_height=3.91e-10/2, std_range=1, demo=False)
Step 1: Height = 1.95e-10, Width = 9.41e-08, Miscut = 0.119°
Step 2: Height = 1.95e-10, Width = 9.42e-08, Miscut = 0.119°
Step 3: Height = 1.95e-10, Width = 2.48e-07, Miscut = 0.045°
Step 4: Height = 1.95e-10, Width = 1.30e-07, Miscut = 0.086°
Step 5: Height = 1.95e-10, Width = 1.03e-07, Miscut = 0.109°
Step 6: Height = 1.95e-10, Width = 1.40e-07, Miscut = 0.080°
Step 7: Height = 1.95e-10, Width = 1.09e-07, Miscut = 0.103°
Step 8: Height = 1.95e-10, Width = 1.50e-07, Miscut = 0.075°
Step 9: Height = 1.95e-10, Width = 1.44e-07, Miscut = 0.078°
Step 10: Height = 1.95e-10, Width = 1.47e-07, Miscut = 0.076°
Step 11: Height = 1.95e-10, Width = 1.51e-07, Miscut = 0.074°
Step 12: Height = 1.95e-10, Width = 1.64e-07, Miscut = 0.068°
Step 13: Height = 1.95e-10, Width = 1.29e-07, Miscut = 0.087°
Results:
Average step height = 1.95e-10, Standard deviation = 0.00e+00
Average step width = 1.39e-07, Standard deviation = 3.84e-08
Average miscut = 0.086°, Standard deviation = 0.020°
Step height = 1.95e-10 +- 0.00e+00
Step width = 1.62e-07 +- 8.27e-08
Miscut = 0.090° +- 0.135°
# calculate the height from afm figure
step_heights, step_widths, miscut = analyzer.calculate_simple(x, z, peak_indices, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=2, prominence=1e-13, width=2, style='simple', std_range=1, demo=False)
Step 1: Height = 1.17e-10, Width = 9.41e-08, Miscut = 0.071°
Step 2: Height = -7.96e-11, Width = 9.42e-08, Miscut = -0.048°
Step 3: Height = 3.26e-11, Width = 2.48e-07, Miscut = 0.008°
Step 4: Height = -1.59e-10, Width = 1.30e-07, Miscut = -0.070°
Step 5: Height = 3.95e-10, Width = 1.03e-07, Miscut = 0.221°
Step 6: Height = -2.97e-10, Width = 1.40e-07, Miscut = -0.121°
Step 7: Height = 1.14e-10, Width = 1.09e-07, Miscut = 0.060°
Step 8: Height = 1.53e-10, Width = 1.50e-07, Miscut = 0.058°
Step 9: Height = -3.56e-10, Width = 1.44e-07, Miscut = -0.141°
Step 10: Height = -1.48e-12, Width = 1.47e-07, Miscut = -0.001°
Step 11: Height = -4.59e-11, Width = 1.51e-07, Miscut = -0.017°
Step 12: Height = -3.21e-11, Width = 1.64e-07, Miscut = -0.011°
Step 13: Height = 3.85e-11, Width = 1.29e-07, Miscut = 0.017°
Results:
Average step height = -9.24e-12, Standard deviation = 1.88e-10
Average step width = 1.39e-07, Standard deviation = 3.84e-08
Average miscut = 0.002°, Standard deviation = 0.089°
Step height = 2.92e-12 +- 1.98e-10
Step width = 1.62e-07 +- 8.27e-08
Miscut = 0.020° +- 0.260°
x, z, peak_indices, valley_indices = analyzer.slice_rotate(img_rot, size_rot, j=100, prominence=1e-10, width=1, xz_angle=0, demo=True)
step_heights, step_widths, miscut = analyzer.calculate_fit(x, z, peak_indices, valley_indices, fixed_height=3.91e-10/2, demo=True)
step 0: step_width: 2.91e-09, left_peak: (1.62e-07, 1.37e-10), valley: (2.79e-07, -7.53e-11), right_peak: (2.94e-07, 2.61e-10)
step 1: step_width: 6.61e-10, left_peak: (2.94e-07, 2.61e-10), valley: (3.53e-07, -1.69e-10), right_peak: (4.70e-07, 2.93e-10)
step 2: step_width: 7.06e-10, left_peak: (4.70e-07, 2.93e-10), valley: (5.44e-07, -1.95e-10), right_peak: (5.88e-07, -6.45e-11)
step 3: step_width: 3.13e-10, left_peak: (5.88e-07, -6.45e-11), valley: (6.17e-07, -1.77e-10), right_peak: (6.47e-07, 2.45e-11)
step 4: step_width: 2.24e-10, left_peak: (6.47e-07, 2.45e-11), valley: (6.61e-07, -1.39e-10), right_peak: (7.20e-07, 1.04e-10)
step 5: step_width: 4.08e-10, left_peak: (7.20e-07, 1.04e-10), valley: (7.79e-07, -6.47e-11), right_peak: (8.23e-07, 1.15e-10)
step 6: step_width: 3.86e-10, left_peak: (8.23e-07, 1.15e-10), valley: (8.52e-07, -1.05e-10), right_peak: (9.26e-07, 3.10e-10)
step 7: step_width: 2.25e-09, left_peak: (9.26e-07, 3.10e-10), valley: (1.01e-06, -5.36e-10), right_peak: (1.04e-06, -6.95e-11)
step 8: step_width: 2.22e-10, left_peak: (1.04e-06, -6.95e-11), valley: (1.09e-06, -1.74e-10), right_peak: (1.13e-06, -5.75e-11)
step 11: step_width: 6.30e-09, left_peak: (1.32e-06, 1.54e-10), valley: (1.50e-06, -1.42e-10), right_peak: (1.51e-06, 3.58e-10)
step 12: step_width: 8.73e-10, left_peak: (1.51e-06, 3.58e-10), valley: (1.56e-06, -2.73e-11), right_peak: (1.59e-06, 2.98e-10)
step 13: step_width: 9.07e-10, left_peak: (1.59e-06, 2.98e-10), valley: (1.69e-06, -3.23e-10), right_peak: (1.84e-06, 8.45e-11)
step 14: step_width: 4.62e-10, left_peak: (1.84e-06, 8.45e-11), valley: (1.94e-06, -1.68e-10), right_peak: (2.04e-06, 4.15e-11)
step 15: step_width: 1.42e-09, left_peak: (2.04e-06, 4.15e-11), valley: (2.16e-06, -3.13e-10), right_peak: (2.20e-06, 8.66e-11)
step 16: step_width: 5.52e-09, left_peak: (2.20e-06, 8.66e-11), valley: (2.40e-06, -3.71e-10), right_peak: (2.41e-06, 1.87e-11)
step 17: step_width: 4.86e-10, left_peak: (2.41e-06, 1.87e-11), valley: (2.45e-06, -2.14e-10), right_peak: (2.54e-06, 2.92e-10)
step 18: step_width: 1.52e-09, left_peak: (2.54e-06, 2.92e-10), valley: (2.63e-06, -3.69e-11), right_peak: (2.66e-06, 3.61e-10)
step 19: step_width: 1.19e-09, left_peak: (2.66e-06, 3.61e-10), valley: (2.76e-06, -2.50e-10), right_peak: (2.84e-06, 1.63e-10)
step 20: step_width: 1.03e-09, left_peak: (2.84e-06, 1.63e-10), valley: (2.94e-06, -1.20e-10), right_peak: (2.97e-06, 9.47e-11)
step 21: step_width: 3.41e-10, left_peak: (2.97e-06, 9.47e-11), valley: (3.03e-06, -8.17e-11), right_peak: (3.07e-06, 4.20e-11)
step 22: step_width: 4.06e-10, left_peak: (3.07e-06, 4.20e-11), valley: (3.10e-06, -1.61e-10), right_peak: (3.16e-06, 2.45e-10)
Results:
Average step height = 1.95e-10, Standard deviation = 0.00e+00
Average step width = 1.36e-09, Standard deviation = 1.62e-09
Average miscut = 17.713°, Standard deviation = 11.870°
# with the provided height from literature
step_heights, step_widths, miscut = analyzer.calculate_substrate_properties(img_rot, size_rot, xz_angle=0, prominence=1e-10, width=1,
style='fit', fixed_height=3.91e-10/2, std_range=1, demo=False)
Step height = 1.95e-10 +- 2.58e-26
Step width = 8.41e-10 +- 5.97e-10
Miscut = 18.522° +- 10.322°
The “simple” method with provided step height gives a more accurate results for the step width and miscut:
Step height = 1.95e-10 +- 0.00e+00
Step width = 1.62e-07 +- 8.27e-08
Miscut = 0.090° +- 0.135°
3. X-ray Diffraction and Reciprocal Space Mapping#
3.1 \(2\theta\) -\(\omega\) scans#
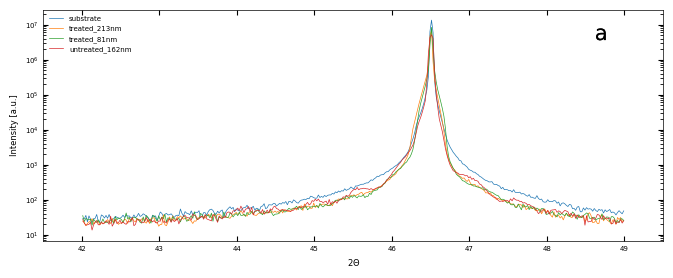
fig, ax = plt.subplots(1, 1, figsize=(8,3))
files = ['./XRD/substrate-XRD_42_49.xrdml', './XRD/treated_213nm-XRD_42_29.xrdml', './XRD/treated_81nm-XRD_42_29.xrdml', './XRD/untreated_162nm-XRD_42_29.xrdml']
labels = ['substrate', 'treated_213nm', 'treated_81nm', 'untreated_162nm']
plot_xrd(ax, files, labels, diff=None, xrange=(41.5, 49.5))
labelfigs(ax, 0, loc='tr', size=15, style='b', inset_fraction=(0.8, 0.1))
3.2 Symmetric Reciprocal Space Mapping (002)#
files = ['./XRD/treated_213nm-RSM_002.xrdml',
'./XRD/treated_81nm-RSM_002.xrdml',
'./XRD/untreated_162nm-RSM_002.xrdml']
titles = ['treated_213nm', 'treated_81nm', 'untreated_162nm']
fig, axes = layout_fig(3, 1, figsize=(6,12))
for i, ax in enumerate(axes):
plot_rsm(ax, files[i], title=titles[i])
labelfigs(ax, i, loc='tr', size=15)
plt.show()
print(f'\033[1mFig.\033[0m reciprocal space mapping in (002) for sample a: treated_213nm, b: treated_81nm, c: untreated_162nm.')
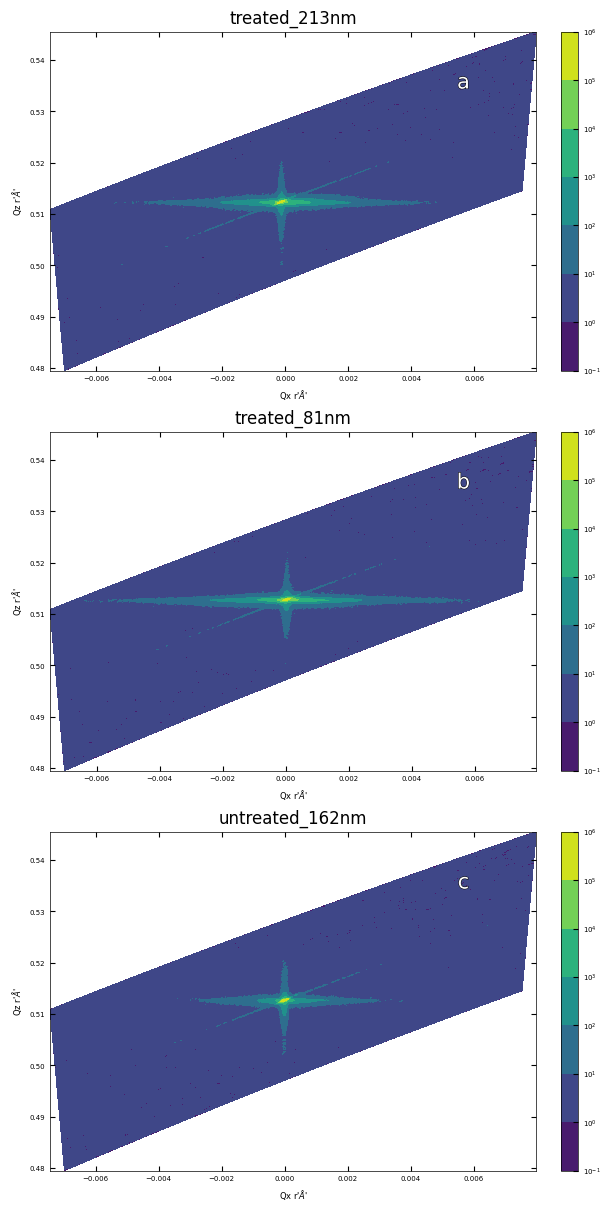
Fig. reciprocal space mapping in (002) for sample a: treated_213nm, b: treated_81nm, c: untreated_162nm.
3.3 Symmetric Reciprocal Space Mapping (103)#
files = ['./XRD/treated_213nm-RSM_103.xrdml',
'./XRD/treated_81nm-RSM_103.xrdml',
'./XRD/untreated_162nm-RSM_103.xrdml']
titles = ['treated_213nm', 'treated_81nm', 'untreated_162nm']
fig, axes = layout_fig(3, 1, figsize=(6,15))
for i, ax in enumerate(axes):
plot_rsm(ax, files[i], title=titles[i])
labelfigs(ax, i, loc='tr', size=15)
plt.show()
print(f'\033[1mFig.\033[0m reciprocal space mapping in (103) for sample a: treated_213nm, b: treated_81nm, c: untreated_162nm.')
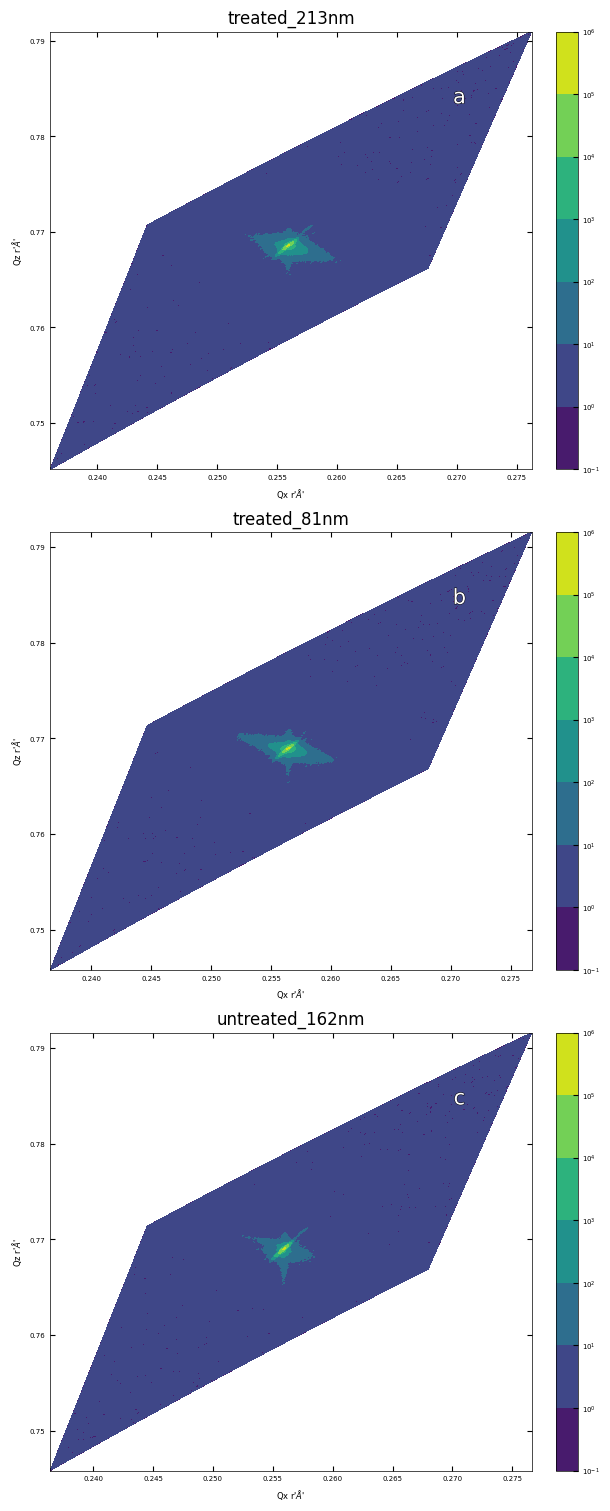
Fig. reciprocal space mapping in (103) for sample a: treated_213nm, b: treated_81nm, c: untreated_162nm.
3.4 Summary Figure of XRD and RSM results#
fig = plt.figure(figsize=(8,10))
ax0 = plt.subplot2grid((4, 2), (0, 0), colspan=2) # colspan=2 means the plot spans 2 columns
files = ['./XRD/substrate-XRD_42_49.xrdml', './XRD/treated_213nm-XRD_42_29.xrdml', './XRD/treated_81nm-XRD_42_29.xrdml', './XRD/untreated_162nm-XRD_42_29.xrdml']
labels = ['substrate', 'treated_213nm', 'treated_81nm', 'untreated_162nm']
plot_xrd(ax0, files, labels, diff=None, xrange=(41.8, 49.2))
labelfigs(ax0, 0, loc='tr', size=15, style='b', inset_fraction=(0.8, 0.1))
files_002 = ['./XRD/treated_213nm-RSM_002.xrdml', './XRD/treated_81nm-RSM_002.xrdml', './XRD/untreated_162nm-RSM_002.xrdml']
for i, file in enumerate(files_002):
ax = plt.subplot2grid((4, 2), (i+1, 0))
plot_rsm(ax, file)
labelfigs(ax, i+1, loc='tr', size=15)
files_103 = ['./XRD/treated_213nm-RSM_103.xrdml', './XRD/treated_81nm-RSM_103.xrdml', './XRD/untreated_162nm-RSM_103.xrdml']
for i, file in enumerate(files_103):
ax = plt.subplot2grid((4, 2), (i+1, 1))
plot_rsm(ax, file)
labelfigs(ax, i+4, loc='tr', size=15)
printing.savefig(fig, 'XRD_RSM')
plt.show()
print(f'\033[1mFig. S2 a\033[0m X-ray Diffraction result for a typical SrTiO3 substrate and samples. \
\033[1mb, c, d\033[0m Reciprocal Space Mapping results in (002) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively. \
\033[1me, f, g\033[0m Reciprocal Space Mapping results in (103) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively.')
Figures/2.AFM_XRD/XRD_RSM.png
Figures/2.AFM_XRD/XRD_RSM.svg
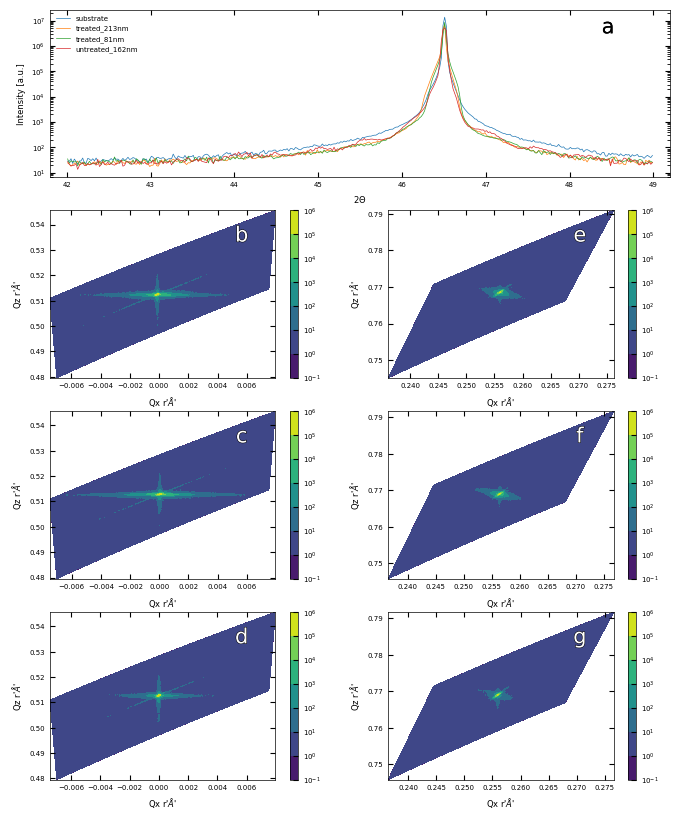
Fig. S2 a X-ray Diffraction result for a typical SrTiO3 substrate and samples. b, c, d Reciprocal Space Mapping results in (002) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively. e, f, g Reciprocal Space Mapping results in (103) orientaion for sample treated_213nm, treated_81nm and untreated_162nm, respectively.


